Global warming due to anthropogenic greenhouse gas emissions poses an immense threat to Earth’s oceans, which serve as a vital climate regulation system. The influx of large quantities of freshwater from melting Arctic sea ice has the potential to critically alter ocean circulation in the North Atlantic. Changes to the physical properties of seawater in the North Atlantic could eventually lead to the collapse of the Atlantic Meridional Overturning Circulation (AMOC), an event that would result in potentially catastrophic changes to climate in the Northern Hemisphere. Predictive climate models have noted that this shift could occur in the future, developing a series of warnings that could help understand more accurately when this major climate shift could occur. Writing in Science Advances, Van Westen and colleagues report the findings of the Community Earth System Model (CESM) and their predictions regarding the impacts of the AMOC’s collapse.
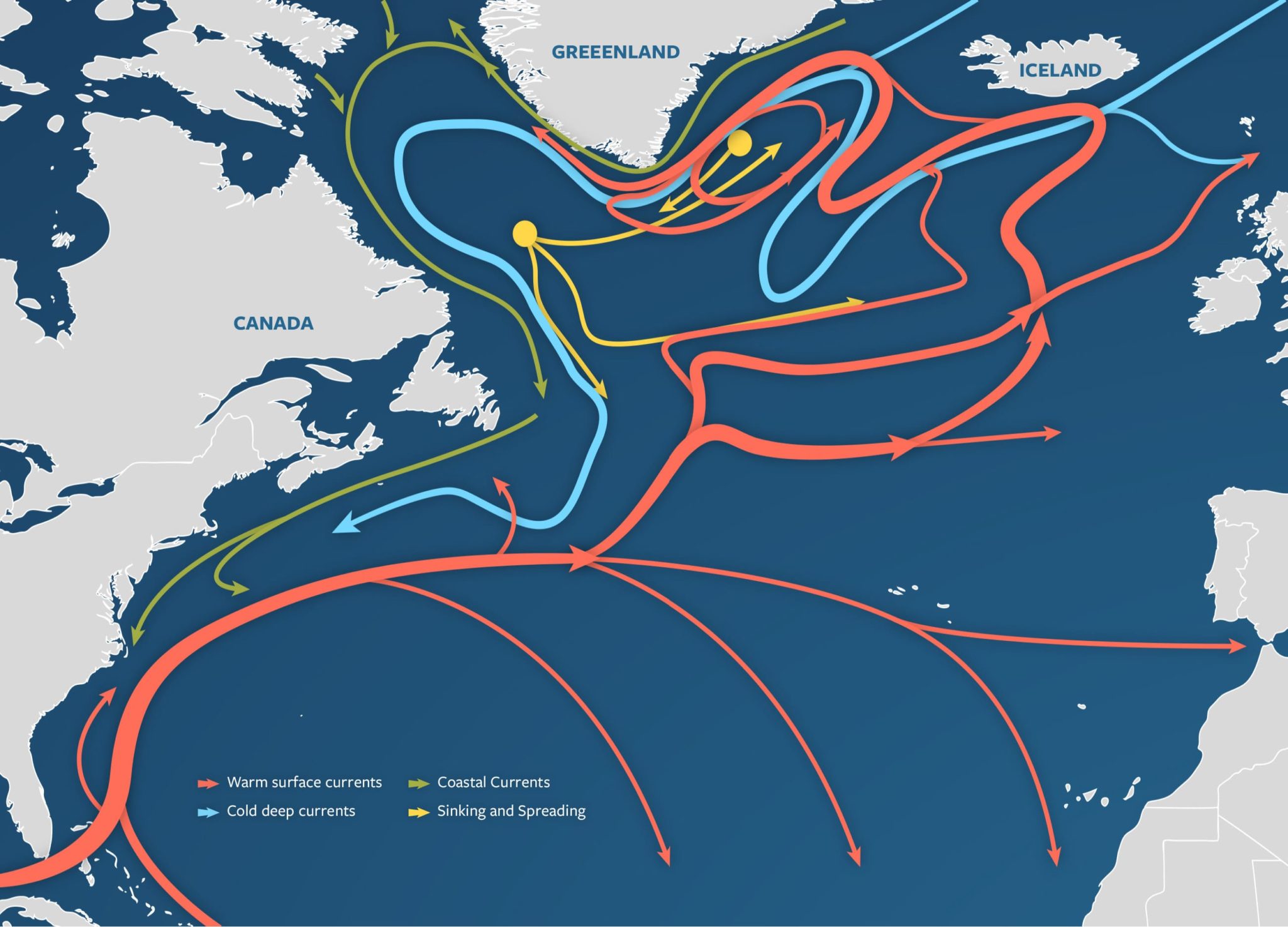 Figure 1: A visualization of the Atlantic Meridional Overturning Circulation (Adapted from “The Ocean Conveyor – Woods Hole Oceanographic Institution,” n.d.).
Figure 1: A visualization of the Atlantic Meridional Overturning Circulation (Adapted from “The Ocean Conveyor – Woods Hole Oceanographic Institution,” n.d.).
The AMOC is a “tipping element” of Earth’s climate, meaning that it is very sensitive to changes in salinity and temperature and could have substantial, reverberating climate impacts if disrupted (Armstrong et al., 2022). Since 1950, oceanographic and climate data have displayed that AMOC strength has significantly decreased and is potentially in its weakest state over the past thousand years (Caesar et al., 2021). These changes largely result from an increased freshwater flux into the North Atlantic due to high rates of Arctic sea ice melting as a product of anthropogenic climate warming. This methodical increase in freshwater flux into the North Atlantic could eventually lead to the collapse of this critical ocean circulation system, an event that would have severe impacts on temperature and weather patterns, especially in the Northern Hemisphere. Prior predictive climate models, which fail to encapsulate Earth climate systems as accurately as the model used by Van Westen and colleagues, have not yet modeled an AMOC collapse. Van Westen et al.’s 2024 study is the first to definitively model this crucial climate tipping point.
Van Westen and colleagues performed their study in CESM version 1.0.5, a complex climate model that simulates earth systems (Danabasoglu et al., 2020). The research team set a preindustrial control simulation with corresponding earth and ocean system conditions at model year 0. To model sea ice melt, they added a methodical, yet variable freshwater flux from the Arctic into the North Atlantic which was increased linearly through model year 2200. This gradual increase in freshwater flux into the North Atlantic corresponded to a gradual decrease in AMOC strength, consistent with predictions made by the research team. AMOC strength began diminishing in model year 800 and abruptly collapsed in model year 1758. This collapse represented a five-fold decrease in AMOC strength over the course of a century from model years 1750 to 1850, a shockingly abrupt change given the slow, consistent freshwater flux into the North Atlantic. By model year 2000, northward heat transport by the AMOC in the Atlantic decreased to nearly zero.
 Figure 2: AMOC strength at 1000m depth and 26° N latitude. Yellow band shows the range of previously observed AMOC strength (Adapted from Van Westen et al., 2024).
Figure 2: AMOC strength at 1000m depth and 26° N latitude. Yellow band shows the range of previously observed AMOC strength (Adapted from Van Westen et al., 2024).
Researchers found influential and dynamic changes to physical properties in oceans across the globe with AMOC collapse. Sea surface temperatures (SST) in the Northern Hemisphere after AMOC collapse significantly cooled, with differences as large as 10℃ observed off the coast of western Europe. SSTs increased slightly in the Southern Hemisphere due to the near absence of northward heat transport by the AMOC. Dramatic shifts in salinity in the upper 100 meters of the ocean were observed in addition to the complete interruption of deep ocean convection in the North Atlantic. Sea-level also rose nearly 70 cm in some regions of the coastal Atlantic due to AMOC collapse.
Researchers also investigated potential effects of AMOC collapse on climate and sea-ice extent in both the Northern and Southern Hemispheres. Significant changes to Hadley Cell air circulation and the subtropical jet stream were observed. Sea ice coverage in the Arctic extended to 50°N in the Arctic (current sea ice rarely forms below 60°N), while Antarctic sea-ice retreated. Model outputs showed atmospheric temperature decreases by around 3℃ per decade in the Northern Hemisphere, a rate at which human adaptation efforts would be largely impossible (current rates of temperature increase due to anthropogenic climate warming are ~0.2℃). These temperature shifts were amplified by ice-albedo feedback, where increased ice coverage in the Northern Hemisphere after AMOC collapse reflects a larger amount of solar radiation back into space, reducing atmospheric temperatures further. Additionally, precipitation patterns in tropical regions shifted with a slight increase in atmospheric temperature in the Southern Hemisphere after AMOC tipping. These results explicitly demonstrate that AMOC tipping would have dramatic, cascading climate impacts across the globe.
Van Westen and colleagues’ study was also the first of its kind to develop a comprehensive warning system for AMOC collapse based on historical climate and oceanographic data and model predictions. Observation of freshwater transport at 34°S, an important proxy for AMOC strength, and the identification of a minimum value for freshwater transport at which AMOC collapse could occur are essential characteristics of AMOC tipping that Van Westen and colleagues identified. These markers of AMOC strength provide an observable set of characteristics that could help predict AMOC collapse in real life.
This research is especially unique because it provides a definitive, model-based answer to the question of whether AMOC collapse can occur in climate models. Prior researchers assumed that AMOC tipping was highly theoretical and would not be predicted in a model that accurately accounts for complicated elements of climate systems. Van Westen and colleagues’ findings clearly demonstrate that AMOC tipping is not only possible, but highly likely under sufficient freshwater influx due to melting Arctic ice.
While the simulation performed by Van Westen et al. (2024) represents an effective predictor of major changes in Atlantic circulation, more data is needed to optimize predictive climate models and apply findings to real climate systems. Van Westen’s research team was unable to provide a meaningful estimate of when an actual AMOC tipping event could occur due to uncertainties in the rate and effects of future climate change. In a paper examining crucial climate tipping points, another European research team estimated that AMOC collapse could occur anywhere from 15-300 years from now, with researchers agreeing that collapse may most likely occur 50 years from now (Armstrong et al., 2022). Another study by researchers from the University of Copenhagen predicted with 95% confidence that tipping may occur from 2025-2095 (Ditlevsen & Ditlevsen, 2023). Precise monitoring of the physical changes in the North Atlantic and stringent data collection are essential to develop more accurate predictions of when AMOC collapse could occur in real life.
This research by Van Westen and colleagues shows evidence that the AMOC could reach a tipping point due to freshwater transport, temperature changes, and salinity changes in the Atlantic, leading to catastrophic climate impacts across the globe, especially in the Northern Hemisphere. Predictive models of major climate events are instrumental in helping communicate the severity of anthropogenic climate change and should be utilized by scientists, policymakers, and advocates throughout the transition away from our reliance on high emission fossil fuel combustion.
References:
Armstrong McKay, D. I., Staal, A., Abrams, J. F., Winkelmann, R., Sakschewski, B., Loriani, S., Fetzer, I., Cornell, S. E., Rockström, J., & Lenton, T. M. (2022). Exceeding 1.5°C global warming could trigger multiple climate tipping points. Science, 377(6611), eabn7950. https://doi.org/10.1126/science.abn7950
Caesar, L., McCarthy, G. D., Thornalley, D. J. R., Cahill, N., & Rahmstorf, S. (2021). Current Atlantic Meridional Overturning Circulation weakest in last millennium. Nature Geoscience, 14(3), 118–120. https://doi.org/10.1038/s41561-021-00699-z
Danabasoglu, G., Lamarque, J.-F., Bacmeister, J., Bailey, D. A., DuVivier, A. K., Edwards, J., Emmons, L. K., Fasullo, J., Garcia, R., Gettelman, A., Hannay, C., Holland, M. M., Large, G., Lauritzen, P. H., Lawrence, D. M., Lenaerts, J. T. M., Lindsay, K., Lipscomb, W. H., Mills, M. J., … Strand, W. G. (2020). The Community Earth System Model Version 2 (CESM2). Journal of Advances in Modeling Earth Systems, 12(2), e2019MS001916. https://doi.org/10.1029/2019MS001916
Ditlevsen, P., & Ditlevsen, S. (2023). Warning of a forthcoming collapse of the Atlantic meridional overturning circulation. Nature Communications, 14(1), 4254. https://doi.org/10.1038/s41467-023-39810-w
The Ocean Conveyor—Woods Hole Oceanographic Institution. (n.d.). https://www.whoi.edu/. Retrieved April 21, 2024, from https://www.whoi.edu/knowyourocean/oceantopics/how-the-ocean-works/ocean-circulation/the-ocean-conveyor/
Van Westen, R. M., Kliphuis, M., & Dijkstra, H. A. (2024). Physics-based early warning signal shows that AMOC is on tipping course. Science Advances, 10(6), eadk1189. https://doi.org/10.1126/sciadv.adk1189