Recent progress has been made in malaria elimination using insecticides, bed nets, and clinical management (CDC). However, efficient mosquitos carrying deadly species of malaria parasites paired with favorable climate in regions where the disease is prevalent, weak infrastructure, and high intervention costs create difficult barriers to malaria elimination (CDC). Similarly, new threats are arising from drug- and insecticide-resistant parasites that pose a risk of expansion to densely populated areas with no prior exposure to the parasites and an unprepared public health system (Garrood et al., 2022). Amidst malaria control efforts, many scientists are investigating a promising new method of inhibiting mosquitos’ ability to transmit malaria: gene drives.
Gene drives, or “selfish” genes, are transmitted at a frequency greater than 50%, above what is predicted by Mendelian genetics. These genes occur naturally in populations; however, we can also engineer gene drives using CRISPR/Cas9 technology to insert a desired gene into a population (Bier, 2022, Hillary et al., 2020). The CRISPR/Cas9 system targets a certain DNA sequence and cuts it, after which the DNA will repair itself either randomly (end-joining, or EJ) or following a provided template (homology-directed repair, or HDR) (Adolfi et al., 2020, Hillary et al., 2020). In the case of gene drives, researchers create a drive, or set of genes, containing the desired genes to be inserted as well as genes encoding necessary CRISPR/Cas9 elements. The presence of CRISPR/Cas9 elements ensures that offspring can make the CRISPR/Cas9 proteins necessary to insert the desired gene into their genome. The desired inserted genes can serve a variety of purposes; in the case of malaria, scientists are inserting antimalarial effector genes into gene drives (Gantz et al, 2015, Isaacs et al., 2012).
Prior research demonstrates use of a gene drive, nRec, to insert antimalarial effector genes that inhibit parasite transmission into a mosquito population (Gantz et al, 2015, Isaacs et al., 2012). The drive targets the essential kynurenine hydroxylase (kh) gene, causing a loss-of-function mutation that reduces female survival and reproduction (Gantz et al., 2015). This type of drive, although effective in preventing malaria transmission, ultimately proves unsustainable given that the loss-of-function results in poor fitness and selection against the drive. A recent study proposes a solution to this problem via a gene drive rescue system that restores the function of the kh gene (Adolfi et al., 2020). The restoration of kh gene function ensures sustainable propagation of the drive and alleviates the fitness cost of the drive.
In a study led by Adriana Adolfi at University of California, Irvine, researchers aimed to target the same essential kh gene while also providing a rescue sequence that maintains the function of the gene (Adolfi et al. 2015). Although their drive does not include the antimalarial effector genes used in prior drives, they do not anticipate any challenges to incorporating the genes. Their approach addresses two problems: it minimizes the impact of drive-resistant alleles and reduces the fitness cost of the drive. By targeting the essential kh gene, drive-resistant mosquitoes are selected out because of the fitness cost that they take on while those that are not resistant do not suffer a fitness cost. The EJ repair that occurs in drive-resistant mosquitoes results in a loss-of-function mutation to the kh gene that makes the mosquitoes less fit for survival than their nonresistant counterparts who repair the Cas9 cleavage with HDR. HDR restores the original sequence of the kh gene by repairing the DNA according to a provided template, thus preventing the loss-of-function mutation, the fitness cost, and ultimately a population crash. In contrast, if the researchers had chosen to target a non-essential gene, this undesirable EJ event would not confer a fitness cost and would then be maintained while preventing transmission of the drive. Ultimately, the drive acts by replacing the previously targeted portion of the kh gene with the original kh sequence to alleviate prior fitness costs of the drive and minimize survival of drive-resistant individuals.
Reckh mosquitos, those that acquired the drive, demonstrated fitness equal to wild-type (WT) mosquitos, while mosquitos with a loss-of-function mutation in the kh gene demonstrated decreased fitness and were eliminated from the population. The drive was released into caged mosquito populations with varied ratios of Reckh males to WT males (1:1, 1:3, and 1:9). In all cages except for one 1:3 cage, the drive was present in more than 95% of mosquitoes by the eleventh generation. Notably, the population did not crash, and the drive was not selected out of the population. In prior research with the nRec drive, few cages reached greater than 95% introduction of the drive before the population crashed or the drive was selected out. The consistent spread of the Reckh drive demonstrates its ability to combat previous challenges of drive sustainability.
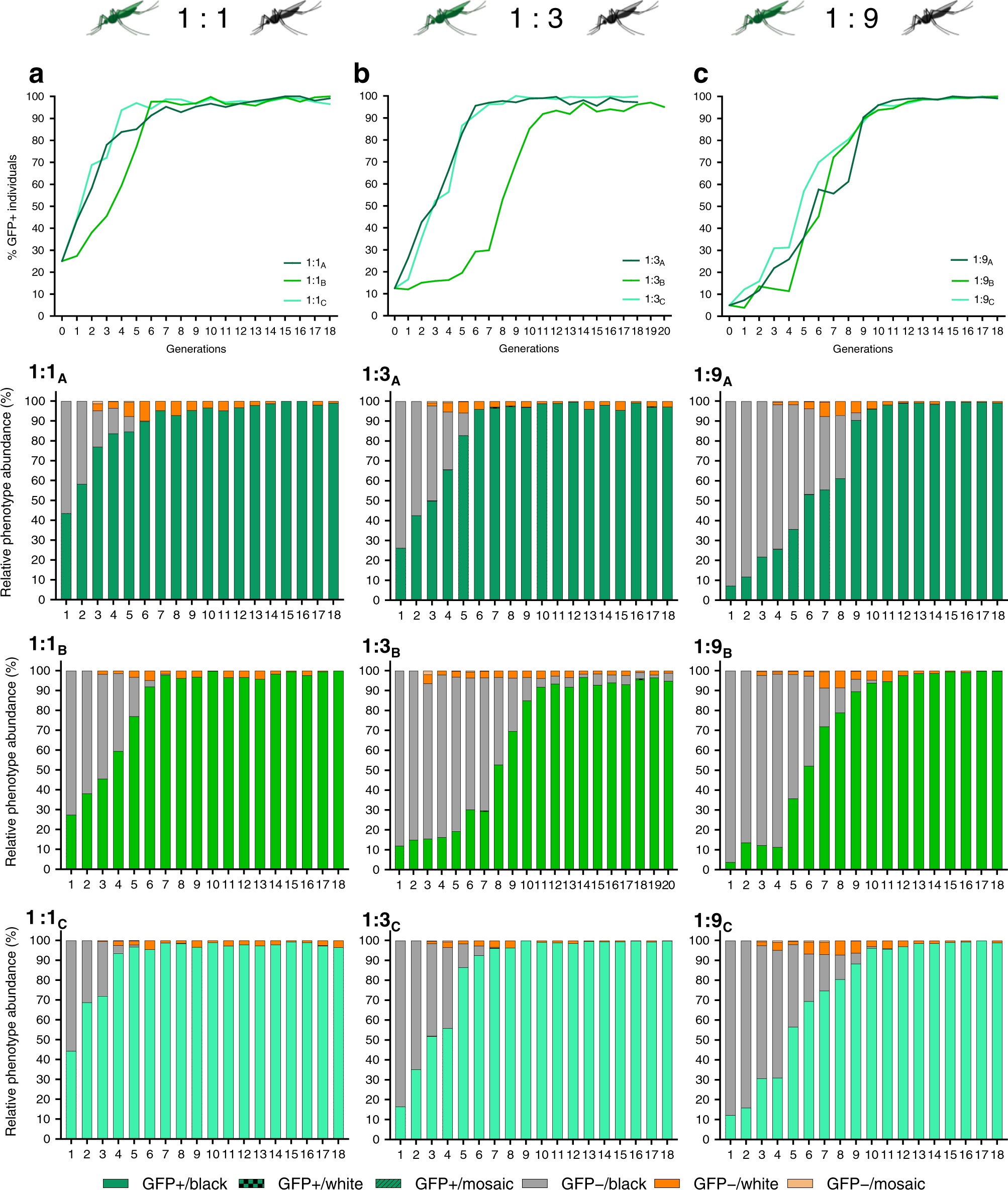
Figure 1: The drive reached >95% introduction by the eleventh generation in populations seeded with ratios of Reckh:WT males of 1:1, 1:3, and 1:9 while drive-resistant individuals were eliminated from the population. Green represents individuals with the drive, gray represents those without the drive, and orange represents drive-resistant individuals. (Adapted from Adolfi et al., 2020)
Furthermore, analysis of mosquitos with non-functional kh– alleles revealed that they were selected out of the population at a rate higher than expected. The researchers propose that lethal/sterile mosaicism factors into elimination of individuals with non-functional kh– alleles. During reproduction, offspring inherit one kh allele from their mother and one kh allele from their father. In the proposed model, CRISPR/Cas9 elements inherited from a mother with a non-functional kh– allele target and cut the WT kh+ paternal allele. This results in EJ mutations in the WT paternal allele that render the offspring less fit and accelerates its elimination from the population. The recoded kh gene sequence is not present in the mothers because they have a non-functional kh– allele, thus the mosquitoes cannot use HDR to repair the cut paternal allele and must instead use EJ. This lethal/sterile mosaicism ensures that non-functional kh– alleles act dominantly to eliminate drive-resistant individuals from the population.
The proposed gene drive shows promise towards sustainability of antimalarial gene drives in caged trials. To further explore the potential of this gene drive, the researchers propose further experimentation with the antimalarial effector genes inserted into their drive. Based on prior research, they do not anticipate any fitness disadvantage to the insertion of the antimalarial genes nor any challenges due to the large size of the drive. Although much research is required before this gene drive can be implemented in wild populations, it introduces a sustainable and effective method of malaria control.
References
- Adolfi, A., Gantz, V. M., Jasinskiene, N., Lee, H.-F., Hwang, K., Terradas, G., Bulger, E. A., Ramaiah, A., Bennett, J. B., Emerson, J. J., Marshall, J. M., Bier, E., & James, A. A. (2020). Efficient population modification gene-drive rescue system in the malaria mosquito Anopheles stephensi. Nature Communications, 11(1), 5553. https://doi.org/10.1038/s41467-020-19426-0
- Bier, E. (2022). Gene drives gaining speed. Nature Reviews Genetics, 23(1), 5–22. https://doi.org/10.1038/s41576-021-00386-0
- Gantz, V. M., Jasinskiene, N., Tatarenkova, O., Fazekas, A., Macias, V. M., Bier, E., & James, A. (2015). Highly efficient Cas9-mediated gene drive for population modification of the malaria vector mosquito Anopheles stephensi. Proceedings of the National Academy of Sciences of the United States of America, 112(49), E6736–E6743. https://doi.org/10.1073/pnas.1521077112
- Garrood, W. T., Cuber, P., Willis, K., Bernardini, F., Page, N. M., & Haghighat-Khah, R. E. (2022). Driving down malaria transmission with engineered gene drives. Frontiers in Genetics, 13, 891218. https://doi.org/10.3389/fgene.2022.891218
- Hillary, V. E., Ceasar, S. A., & Ignacimuthu, S. (2020). Chapter 18 – Genome engineering in insects: Focus on the CRISPR/Cas9 system. In V. Singh & P. K. Dhar (Eds.), Genome Engineering via CRISPR-Cas9 System (pp. 219–249). Academic Press. https://doi.org/10.1016/B978-0-12-818140-9.00018-0
- How Can Malaria Cases and Deaths Be Reduced? (2019, January 28). Centers for Disease Control and Prevention. https://www.cdc.gov/malaria/malaria_worldwide/reduction/index.html
- Isaacs, A. T., Jasinskiene, N., Tretiakov, M., Thiery, I., Zettor, A., Bourgouin, C., & James, A. A. (2012). Transgenic Anopheles stephensi coexpressing single-chain antibodies resist Plasmodium falciparum development. Proceedings of the National Academy of Sciences of the United States of America, 109(28), E1922–E1930. https://doi.org/10.1073/pnas.1207738109