Saffron is a popular food condiment known to have a positive effect against neurodegenerative diseases such as dementia, Alzheimer’s, and Parkinson’s. Saffron could potentially alleviate some of the biological hallmarks of Alzheimer’s disease, including the accumulation of Aβ plaques outside of cells and neurofibrillary tangles (NFTs) inside of cells. It could also target the neurotransmitter system that involves acetylcholine (ACh), which when disrupted can lead to the progression of dementia. This system, called the cholinergic system, is essential for attention, learning, and memory. ACh plays a role in memory acquisition, encoding, and consolidation (Patel et al. 2024).
A recent study looked at how saffron extract (CSE) supplementation affects behavioral activity, memory, Aβ plaques, and NFTs in rats. The study also investigated how saffron affects a disrupted cholinergic system. When the system breaks down, there is an increase in Aβ plaques and NFTs. There is also an increase in AChE, which is the protein that breaks down ACh. All of these factors can ultimately contribute to Alzheimer’s. The current study investigated how CSE could reduce these effects and ultimately prevent progression of dementia-related diseases (Patel et al. 2024).
In order to study the effects of CSE, the researchers used the drug scopolamine to disrupt the cholinergic system in rats. Scopolamine mimics the memory loss observed in dementia, and it is often used to study the cholinergic system in animal models of dementia. It primarily impairs the acquisition and encoding of memories (Paul 2019). Scopolamine increases the level of AChE in the brain, which means that more ACh is broken down. With less ACh available, the cholinergic system is disrupted, causing cognitive impairments (Figure 1). The study observed the potential of CSE to reduce this disturbance. They used behavioral tests to observe memory and spatial acquisition. Blood samples and brain samples were collected for examination (Patel et al. 2024).

Figure 1. The role of acetylcholinesterase (AChE). AChE breaks down acetylcholine (ACh) as it moves from one neuron to another. An increase in AChE leads to a decrease in ACh. (Adapted from What is the role of acetylcholinesterase at a synapse?)
The researchers found that CSE supplementation significantly improved behavioral activity and memory in rats. CSE improved performance on the reversal task, which requires the ability to switch between learned and new responses, a reflection of cognitive flexibility.
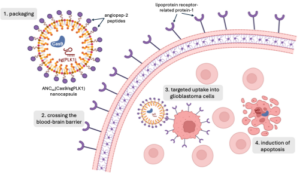
Importantly, the study offers insight into the biological parameters that lead to the improved cognitive abilities. CSE lowered the levels of AChE, meaning that less acetylcholine was broken down. The dose of 20 mg/kg of CSE lowered the amount of AChE in the scopolamine-treated rats as significantly as Rivastigmine did (Figure 2). Rivastigmine is a pharmaceutical drug currently used to treat symptoms of Alzheimer’s by inhibiting AChE (Emre et al. 2010). An increase in the level of acetylcholine can improve memory, as demonstrated by the improved performance of the rats on the memory tests. ACh has been shown to promote nerve conduction related to memory and learning ability, helping memories get encoded and consolidated in the brain.

Figure 2. CSE supplementation lowered the levels of AChE and inhibited its effects in rats who received scopolamine administration. Thus, less acetylcholine was broken down and disruption of the cholinergic system was attenuated. This can lead to an improvement in memory (Adapted from Patel et al, 2024).
CSE supplementation also significantly reduced the formation of Aβ plaques and NFTs, the two classical pathological hallmarks of Alzheimer’s in the hippocampus in the brain. The 20 mg/kg dose of CSE produced the most significant reduction. CSE reduced the number of Aβ plaques and NFTs more than Rivastigmine did. In fact, the Rivastigmine group still had significant levels of NFTs and Aβ plaques (Figure 3). Accumulation of Aβ plaques and NFTs lead to neuronal dysfunction and ultimately dementia. Moreover, AChE and Aβ can bind together to form complexes that exaggerate neurodegeneration even more. Thus, if CSE can attenuate their formation, neuronal function may be preserved, preventing the progression of dementia.

Figure 3. CSE supplementation lowered the density of Aβ plaques in rats who received scopolamine administration. This can lead to an improvement in memory (Adapted from Patel et al, 2024).

Figure 4. Summary of the article. Scopolamine administration led to a disruption of the cholinergic system (a part of the nervous system involving acetylcholine that helps with memory and learning), neuroinflammation, and accumulation of NFT and Aβ plaques. When CSE (saffron extract) was administered, the rats’ memory improved and the biomarkers of Alzheimer’s were reduced (Adapted from Patel et al, 2024).
Because CSE supplementation reduces the formation of Aβ plaque and NFTs in the hippocampus, inhibits the activity of the enzyme that breaks down acetylcholine, and counters neuroinflammation, a saffron-based botanical supplement could be used to help manage dementia. Currently, manufactured drugs such as donepezil and memantine have been used to treat symptoms of Alzheimer’s, but saffron could potentially offer a safer alternative that is just as effective (D’Onofrio et al. 2021). The current study tested the effects of CSE against Rivastigmine, another drug that is approved to treat symptoms of Alzheimer’s. CSE produced the same improvements as Rivastigmine. Rivastigmine, along with other chemical drugs that inhibit AChE in order to treat Alzheimer’s, has been shown to cause gastrointestinal side effects and does not have great patient compliance (Emre et al. 2010). Saffron supplementation could offer an effective treatment without such side effects, increasing quality of life. The research offers promise that saffron can improve the cognitive impairments associated with Alzheimer’s and dementia.
Literature Cited
Emre M, Bernabei R, Blesa R, Bullock R, Cunha L, Daniëls H, Dziadulewicz E, Förstl H, Frölich L, Gabryelewicz T, et al. 2010. Drug Profile: Transdermal Rivastigmine Patch in the Treatment of Alzheimer Disease. CNS Neurosci Ther. 16(4):246–253.
D’Onofrio G, Nabavi SM, Sancarlo D, Greco A, Pieretti S. 2021. Crocus Sativus L. (Saffron) in Alzheimer’s Disease Treatment: Bioactive Effects on Cognitive Impairment. Curr Neuropharmacol. 19(9):1606–1616.
Patel KS, Dharamsi A, Priya M, Jain S, Mandal V, Girme A, Modi SJ, Hingorani L. 2024a. Saffron (Crocus sativus L.) extract attenuates chronic scopolamine-induced cognitive impairment, amyloid beta, and neurofibrillary tangles accumulation in rats. Journal of Ethnopharmacology. 326:117898.
Paul J. 2019. Chapter 5 – Experimental Medicine Approaches in CNS Drug Development. In: Nomikos GG, Feltner DE, editors. Handbook of Behavioral Neuroscience. Vol. 29. Elsevier. (Translational Medicine in CNS Drug Development). p. 63–80. [accessed 2024 Feb 25].
What is the role of acetylcholinesterase at a synapse? | Socratic. Socratic.org. [accessed 2024 Mar 21].