To solve complex human health issues, scientists have more recently turned to biomimicry. Biomimicry, also known as biomimetics, is a field that develops synthetic materials, systems or machines that are derived from the principles of natural biological processes (Nature). Concepts within biomimetics are currently being used to design regenerative medicine and newer drugs for diseases such as cancer. In fact, within Bowdoin, Professor Benjamin Gorske, is undertaking a comprehensive research on developing methods to create drugs that attempt to inhibit signaling pathways within cancer, or interrupt the signal transduction pathways involved in the development of plaques in Alzheimer disease.
Professor Gorske explains that these aforementioned diseases can be addressed by “controlling the signaling proteins”. By interrupting the signaling process, the underlying issue of these diseases – which are often so diverse and hard to target – doesn’t need to be treated. However, a difficulty that comes with targeting signaling proteins is that they bind to other molecules over a vast space, and are implicated in many other signaling pathways. Therefore, these proteins cannot simply be targeted by small molecular drugs, as they would be impossible to effectively block the proteins and thus are called “undruggable molecules”. Instead, Professor Gorske turns to attempting to create a biological molecule that can mimic the other molecules that bind to the target signaling protein – which are normally much bigger than current drug molecules.
One of the targets that Professor Gorske looks into in his lab are the signaling proteins within the Hippo pathway. The Hippo pathway normally controls the size of organs by regulating cell proliferation (division of cells) and apoptosis (programmed cell death) (Cell Signaling Technology). Ultimately, this pathway controls the expression of certain genes that are involved in the proliferation process. This pathway is involved in cancer when it is disregulated, as it continuously sends a signal to the nucleus to express these genes encoding for cell proliferation, thus leading to the uncontrollable growth of cancer. The Hippo pathway involves a lot of signaling proteins that are solely involved in this signaling pathway. Most of these signaling proteins contain a WW domain – a distinct functional unit of the protein that mediates the interactions these proteins have with other molecules (EMBL-EBI). All the signaling proteins with this specific domain in this pathway generally bind to proteins with polyproline type 2 helices (PPII). Professor Gorske’s lab aims to design a molecule to contain this PPII structural component to effectively target the signaling proteins (with the WW domain) in the Hippo pathway.
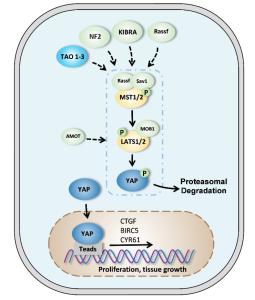
Fig 1: Diagram of the Hippo Pathway. The signaling protein YAP (yes-associated protein/transcription coactivator) is an example of a molecule with the WW domain.
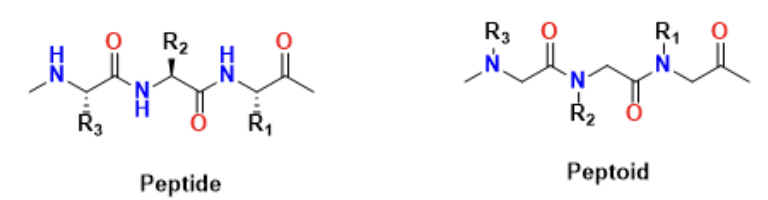
However, we can’t utilize naturally formed proteins with these PPII helices as the proteases in our body would find the related drugs as foreign and destroy them. Thus these helices need to be made artificially, forming molecules called peptoids. Peptoids are a class of molecules that mimic the structure and function of helical peptides (Chongsiriwatana et al., 2008). They are very biostable, relatively easy to make, and most importantly proteases don’t destroy these molecules. Thus Professor Gorske has chosen to make specific peptoids that perhaps could target these signaling proteins in his lab.
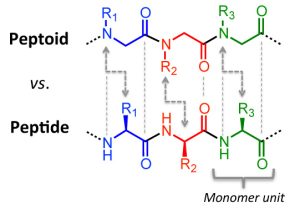
Fig 2: Comparison of the general molecular structure of a naturally-derived peptide versus an artificially-produced peptoid.
However, an issue that arose with creating these peptoids is that the peptoids often folded to form polyproline type 1 helices (PPI) rather than PPII helices. This is due to the differences in orientation of the connected amides (subunits of the peptoids). This leads to peptoids with PP1 helices being much more compressed than peptoids with PPII helices. While they do work as good lung surfactants and other antimicrobials, they are not suitable to target signaling proteins.
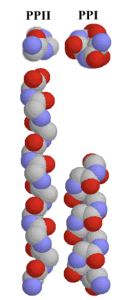
Fig 3: Comparison of the 3D structure of polyproline 1 helix (PPI) and polyproline 2 helix (PPII), and the orientation of the respective amide subunits.
Through experimenting, Professor Gorske has found out in his lab that the best way to encourage amides to connect in the desired PPII orientation is through adding side chains on the amide subunits, and the side chains are additionally thionated – the carbon is double bonded to sulfur instead of oxygen. Currently, he is investigating whether adding more than one thionated residue can lead to the whole peptoid to adopt the desired PPII helical structure.
This method of creating peptoids that Professor Gorske is working to devise can be implicated in so many uses within medicine. As peptoid drugs can mimic biological molecules, they can more precisely target the required proteins, to help inhibit or promote signaling pathways back to normal. Professor Gorske’s research is therefore crucial to the ongoing development of medicine and healthcare.
References
“Biomimetics Articles from across Nature Portfolio.” Nature News, Nature Publishing Group, https://www.nature.com/subjects/biomimetics.
Chen, Yu-An, et al. “WW Domain-Containing Proteins Yap and Taz in the Hippo Pathway as Key Regulators in Stemness Maintenance, Tissue Homeostasis, and Tumorigenesis.” Frontiers in Oncology, vol. 9, 2019, https://doi.org/10.3389/fonc.2019.00060.
Chongsiriwatana, Nathaniel P., et al. “Peptoids That Mimic the Structure, Function, and Mechanism of Helical Antimicrobial Peptides.” Proceedings of the National Academy of Sciences, vol. 105, no. 8, 2008, pp. 2794–2799., https://doi.org/10.1073/pnas.0708254105.
Embl-Ebi. “What Are Protein Domains?” What Are Protein Domains? | Protein Classification, https://www.ebi.ac.uk/training/online/courses/protein-classification-intro-ebi-resources/protein-classification/what-are-protein-domains/.
“Hippo Signaling.” Cell Signaling Technology, 2010, https://www.cellsignal.com/pathways/hippo-signaling-pathway.