The immune system identifies cells in the body by examining molecular stages on the surface of cells, which are known as antigens. Both the human body and mind utilize this information to decide if cells within our body pose a threat to our well-being. Nevertheless, science today continues to struggle with differentiating harmless and harmful cells. For example, cancerous cells may be so similar in antigen identities that they can’t be told apart. However, with a deeper look, researchers may be able to uncover smaller antigens that allows humanity to successfully distinguish between such cells.
Scientists have been using technology that expands cells in uniform across three dimensions to identify such smaller antigens that have previously been unaccounted for. The most recent advancement involved dExPath (decrowding expansion pathology), an improvement with four times greater expansions and stronger resolutions compared to its predecessor, ExPath. dExPath solved many problems by offering multi-round immunostaining, maximized protein epitope (another name for antigens) preservation, and elimination of fluorescent overlaps. Immunostaining normal brain tissue and brain tumor tissue has helped identify numerous improvements and advantages to dExPath. (Valdes et al., 2021)
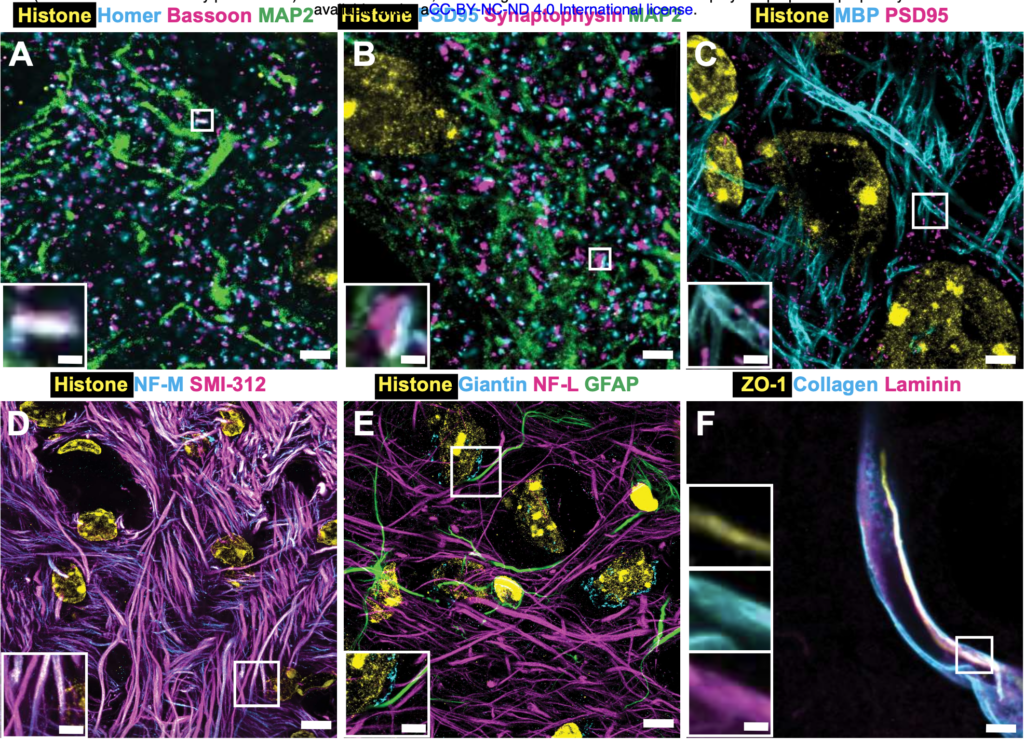
dExPath is a form of microscopy technology that facilitates the immunostaining of cells, so far only applicable to the brain and brain stem cells. Immunostaining utilizes antibodies and conjugated enzymes to generate fluorescence as an indicator of certain antigens and markers on cells of interest. At its core, dExPath is a form of expansion microscopy, a preparation and imaging technique used to visualize biological nanostructures. Wassie et aldescribed the process as synthesizing a dense and even network of swellable polyelectrolyte hydrogel to react with specific nanostructures and physically expand them prior to imaging. Once expanded, immunostaining agents were able to label antigens of interest on cells, all of which were visible from an ordinary laboratory microscope.
Experimental uses of dExPath have allowed researchers to detect distinguishing epitopes and their combinations on glial cell tumors, offering a possibility for the technology’s use in detecting brain cancers. dExPath also facilitated multi-round immunostaining, where multiple forms of staining were implemented to highlight different structures in one image, eliminating the need to run multiple images for each layer of staining.
dExPath has solved many issues that persisted in ExPath. The predecessor failed to preserve some binding sites on protein epitopes, leaving a number of antigens unaccounted for before the use of dExPath. During imaging, the new technology also addressed the prior issue of fluorescent overlaps, particularly due to the presence of lipofuscin, a common waste product among cells. Lipofuscin is often mistaken for a different cellular marker of interest, but decrowding allowed for improved access of antibodies to epitopes, allowing the imaging technology to more clearly recognize its intended targets and ignore the lipofuscin autofluorescence. dExPath’s capacity outperformed Sudan Black B, a stain that reduced the autofluorescence of lipofuscin but weakened other relevant fluorescent signals.
Valdes et al. (2021) used dExPath to expand and investigate human and animal brain cell samples. Samples of normal, low-grade (slow-growth) tumors and high-grade (faster-growth) tumors were examined before and after the expansion process to detect differences in marker visibilities. The samples were immunostained for markers vimetin, Iba1, and GFAP. The presence and frequency of particular markers helped convey new indicators for stages of malignant growth in brain tissues. For example, GFAP and vimentin-labeled low-grade cells were interpreted as an indication of potential future malignancy. Additionally, cells labeled with Iba1 and GFAP indicated a possible increase in tumors with phagocytic (ingestive) properties that made them more invasive.
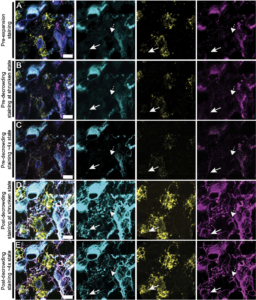
Fluorescence imaged by dExPath is of significantly higher quality and consistency. dExPath revealed that there are significantly more cells of interest, with many corresponding to more different classes of cells, such as macrophages and glioma cells, that are deemed important to glioma pathology than previously expected.
dExPath is already demonstrating tremendous potential in brain disease analysis and diagnosis, as well as accuracy and reliability to an unprecedented degree. The technology’s compatibility with low-cost, conventional microscopes and more commercially available reagents and instruments in basic laboratories makes the technology tremendously accessible and convenient. Its advantages uncovered features after molecular expansion that uncovered cell demographics previously unaccounted for and redefined the aggressive nature of diseased cells. However, there are many more tests that dExPath must undergo, such as assessing its visualization of other neurological diseases or cancers. Continued research concerning dExPath may allow the world of science and medicine to uncover more about mysteries in neurobiology, and possibly introduce dExPath to clinical settings as a new assessment tool for neuropathology diagnosis.
Sources
Valdes, Pablo A., et al. Decrowding Expansion Pathology: Unmasking Previously Invisible Nanostructures and Cells in Intact Human Brain Pathology Specimens. 7 Dec. 2021,
Wassie, Asmamaw T., et al. “Expansion Microscopy: Principles and Uses in Biological Research.” Nature Methods, vol. 16, no. 1, 2019, pp. 33–41, https://doi.org/10.1038/s41592-018-0219-4.
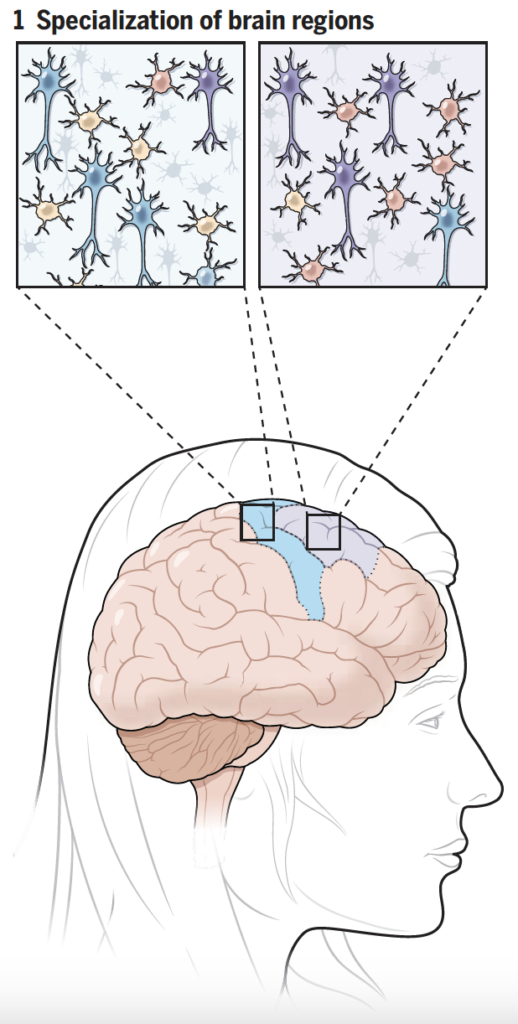
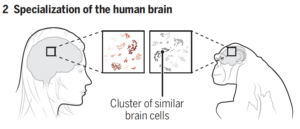
 Comparisons of cell composition in regions across the brain resulted in findings from researchers under Siletti from the University of North Carolina at Chapel Hill and Jorstad from Harvard University. The two groups found that rather than mainly having different types of cells in different parts of the brain, some different parts of the brain shared the same cells but had different proportions of these cells. There were some exceptions, such as inhibitory neurons in the primary visual cortex, although the explanation of this finding is unclear. Such results change the understanding of evolutionary diversity in that diversification does not depend heavily on having many different cell types, but rather on having varying proportions of cells with small differences.
Comparisons of cell composition in regions across the brain resulted in findings from researchers under Siletti from the University of North Carolina at Chapel Hill and Jorstad from Harvard University. The two groups found that rather than mainly having different types of cells in different parts of the brain, some different parts of the brain shared the same cells but had different proportions of these cells. There were some exceptions, such as inhibitory neurons in the primary visual cortex, although the explanation of this finding is unclear. Such results change the understanding of evolutionary diversity in that diversification does not depend heavily on having many different cell types, but rather on having varying proportions of cells with small differences.