In Ancient Egypt, a diet of whole grains was prescribed to patients with frequent urination and emaciation (History of diabetes, 2020). This condition, similarly documented by physicians in Ancient Greece, was likely first called “diabetes” by Apollonius of Memphis in the third century BCE. By the fifth century CE, type 1 diabetes had been differentiated from type 2, and in 1776, English physician Matthew Dobson confirmed the presence of excess glucose in the urine of diabetic patients. Canadian physician Frederick Banting and his colleagues successfully used insulin injections to treat a diabetic patient in 1922; this remains the predominant method of treatment today. Another milestone was reached this past November, when the drug Teplizumab (under the name Tzield) gained FDA approval. Teplizumab showed potential in delaying the onset of clinical type 1 diabetes in adults and pediatric patients 8 years and older who have not yet developed the condition (Commissioner of the FDA, 2022).
Type 1 diabetes, otherwise known as juvenile diabetes or insulin-dependent diabetes, is a form of diabetes mellitus in which a deficiency of insulin causes hyperglycemia (high blood sugar). It is referred to as “juvenile diabetes” because the symptoms of the condition typically appear in adolescence and is the result of an autoimmune disorder. The onset of type 1 diabetes depends on environmental factors that interact with predisposing genes to induce a long-term autoimmune attack against the pancreatic β cells, the insulin-producing cells of the pancreatic islets of Langerhans (de Beeck and Eizrik, 2018).
When left untreated, juvenile diabetes poses serious health risks. The buildup of sorbitol—a sugar alcohol that is manufactured from glucose—within the eye can result in cataracts and blindness (CDC, 2022). Excess sorbitol is also associated with blood vessel lesions and gangrene. Other significant health risks for individuals with juvenile diabetes include ketoacidosis, the buildup of acidic ketones that can cause diabetic comas and diminished brain function, and hypoglycemia, otherwise known as “insulin shock,” resulting from an overdose of insulin or the failure to eat and potentially causing nerve damage and death (CDC, 2022). Approximately 1.6 million Americans live with type 1 diabetes, including 200,000 youth. Furthermore, despite significant developments in the treatment of type 1 diabetes, desired glycemic targets are rarely achieved in patients, who continue to face a higher risk of complications and death because of their condition (Herold et. al., 2019).
In those who are genetically susceptible to type 1 diabetes, there are two asymptomatic stages prior to the development of overt hyperglycemia, the clinical disease which requires insulin treatment. Stage 1 is characterized by the appearance of autoantibodies targeting pancreatic cells and stage 2 involves dysglycemia, an abnormality in blood sugar stability (Herold et. al., 2019). In this case, metabolic responses to high levels of glucose could be impaired, but other metabolic indexes, such as the level of glycosylated hemoglobin, are normal. During these stages, insulin treatment is not required. The goal of Teplizumab is to delay the development of clinical (stage 3) diabetes in those currently in stages 1 and 2 (Herold et. al., 2019).
Teplizumab is an Fc receptor–nonbinding anti-CD3 monoclonal antibody that modifies CD8+ T lymphocytes, which are part of the body’s adaptive immune response and thought to be important effector cells that kill β cells in the pancreas (Herold et. al., 2019). Evidence indicates that type 1 diabetes is initiated by both CD4+ and CD8+ T cells (Li and Qin, 2014). Autoreactive T cells differentiate into effector (CD4+) cells by engaging β-cell antigens on local antigen-presenting cells; these effector CD4+ T cells stimulate other immune cells to target β cells, whereas the cytotoxic CD8+ T cells can directly kill β cells via cell-to-cell contact (Gearty et. al., 2022). The CD3 protein complex is involved in activating both the cytotoxic and helper T-cells (Yang et. al., 2005); thus, the anti-CD3 properties of Teplizumab can inhibit the T cell–mediated damage to β-cells.
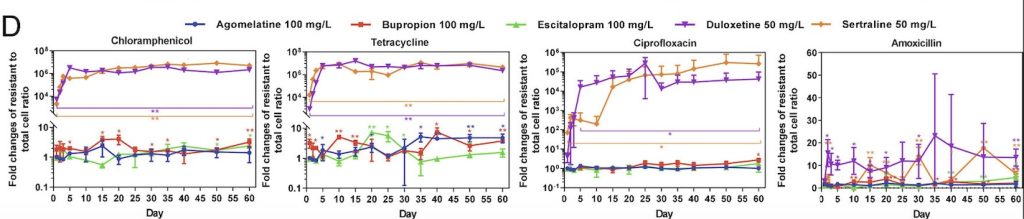
In the Teplizumab trial, which was conducted over a 7-year period and led by Dr. Kevan Herold of Yale University, patients were randomly assigned to a single 14-day course of Teplizumab or placebo. Seventy-two percent of the participants were under the age of 18, and the majority had siblings with clinical type 1 diabetes, meaning they were at a high risk of developing the condition themselves (Herold et. al., 2019). Furthermore, of the 55 patients who were under 18, 47 had a confirmed dysglycemic oral glucose-tolerance test, one of the hallmarks of stage 2 type 1 diabetes, before undergoing randomization in the trial. Follow-up for progression to clinical type 1 diabetes was performed via oral glucose-tolerance tests every 6 months. The study indicated that a 2-week course of treatment with Teplizumab delayed the diagnosis of clinical type 1 diabetes in high-risk participants: following the completion of the trial, 57% of individuals in the Teplizumab trial group were diabetes-free compared to 28% in the placebo group, with a median delay in the diagnosis of clinical diabetes of 2 years (Figure 1). These results also reinforced prior findings that type 1 diabetes is a T-cell mediated condition and showed that immunomodulation before the onset of clinical type 1 diabetes is a promising development in the treatment of this disease (Herold et. al., 2019).
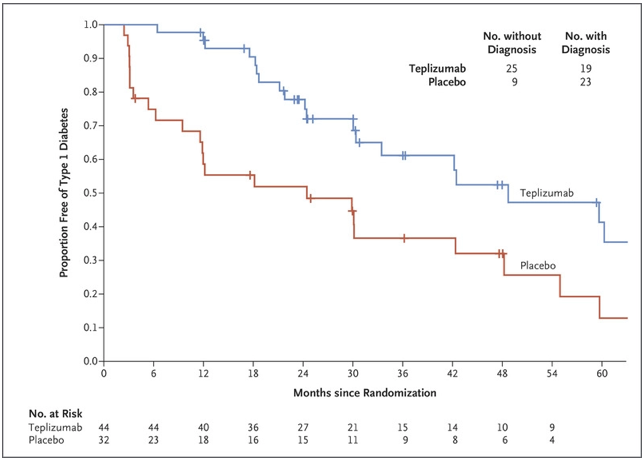
Figure 1. From the time of randomization until the clinical diagnosis of type 1 diabetes, the recipients of the Teplizumab infusion experienced a longer median time until diagnosis than those in the placebo group (adapted from K. C. Herold, et. al.).
Given the prevalence of patients with clinical type 1 diabetes, these results of this study are highly promising and a significant step forward in the mitigation of the harmful side effects of this condition. However, there are several areas which, in an effort to make the benefits of this trial widely applicable, require further research. Since the participants in the study were all relatives of patients with type 1 diabetes, it is currently unknown whether the findings can be applied to those who seem to be at risk for the type 1 diabetes and lack first-degree relatives with the condition (Evans-Molina and Oram, 2023). Furthermore, while patients can be carefully screened for the immunological or metabolic markers of preclinical type 1 diabetes in the research setting, a lack of infrastructure prevents a larger-scale screening of the general public and high-risk populations are typically the only groups surveyed for these symptoms (Evans-Molina and Oram, 2023). Finally, despite the promising results of the newly-approved drug, the current cost of treatment presents a significant barrier regarding access to care: one vial of Tzield costs $13,850, amounting to $193,000 over the 14-day infusion (Rodriguez, n.d.). The exorbitant sum is not surprising, given the high cost of insulin medications and their widely-reported underuse (Herkert et. al., 2019). As a result, further efforts must be taken to ensure that those who are eligible for this new and potentially life-saving medication are able to access it.
Sources:
CDC. (2022, November 3). Prevent diabetes complications. Centers for Disease Control and Prevention. https://www.cdc.gov/diabetes/managing/problems.html
Commissioner, O. of the. (2022, November 18). FDA approves first drug that can delay onset of type 1 diabetes. FDA. https://www.fda.gov/news-events/press-announcements/fda-approves-first-drug-can-delay-onset-type-1-diabetes
de Beeck, A. O., & Eizirik, D. L. (2016). Viral infections in type 1 diabetes mellitus—Why the β cells? Nature Reviews. Endocrinology, 12(5), 263–273. https://doi.org/10.1038/nrendo.2016.30
Dolgin, E. (2023). How a pioneering diabetes drug offers hope for preventing autoimmune disorders. Nature, 614(7948), 404–406. https://doi.org/10.1038/d41586-023-00400-x
Evans-Molina, C., & Oram, R. A. (2023). Teplizumab approval for type 1 diabetes in the USA. The Lancet Diabetes & Endocrinology, 11(2), 76–77. https://doi.org/10.1016/S2213-8587(22)00390-4
Gearty, S. V., Dündar, F., Zumbo, P., Espinosa-Carrasco, G., Shakiba, M., Sanchez-Rivera, F. J., Socci, N. D., Trivedi, P., Lowe, S. W., Lauer, P., Mohibullah, N., Viale, A., DiLorenzo, T. P., Betel, D., & Schietinger, A. (2022). An autoimmune stem-like CD8 T cell population drives type 1 diabetes. Nature, 602(7895), 156–161. https://doi.org/10.1038/s41586-021-04248-x
Herkert, D., Vijayakumar, P., Luo, J., Schwartz, J. I., Rabin, T. L., DeFilippo, E., & Lipska, K. J. (2019). Cost-related insulin underuse among patients with diabetes. JAMA Internal Medicine, 179(1), 112–114. https://doi.org/10.1001/jamainternmed.2018.5008
Herold, K. C., Bundy, B. N., Long, S. A., Bluestone, J. A., DiMeglio, L. A., Dufort, M. J., Gitelman, S. E., Gottlieb, P. A., Krischer, J. P., Linsley, P. S., Marks, J. B., Moore, W., Moran, A., Rodriguez, H., Russell, W. E., Schatz, D., Skyler, J. S., Tsalikian, E., Wherrett, D. K., … Greenbaum, C. J. (2019). An anti-cd3 antibody, teplizumab, in relatives at risk for type 1 diabetes. New England Journal of Medicine, 381(7), 603–613. https://doi.org/10.1056/NEJMoa1902226
History of diabetes: Early science, early treatment, insulin. (2020, June 17). https://www.medicalnewstoday.com/articles/317484
Li, M., Song, L.-J., & Qin, X.-Y. (2014). Advances in the cellular immunological pathogenesis of type 1 diabetes. Journal of Cellular and Molecular Medicine, 18(5), 749–758. https://doi.org/10.1111/jcmm.12270
Masharani, U. B., & Becker, J. (2010). Teplizumab therapy for type 1 diabetes. Expert Opinion on Biological Therapy, 10(3), 459–465. https://doi.org/10.1517/14712591003598843
Rodriguez, A. (n.d.). FDA approves first treatment that delays Type 1 diabetes. Why it could be “game changing.” USA TODAY. Retrieved April 2, 2023, from https://www.usatoday.com/story/news/health/2022/11/18/fda-approves-teplizumab-delays-onset-diabetes/10721707002/
Yang, H., Parkhouse, R. M. E., & Wileman, T. (2005). Monoclonal antibodies that identify the CD3 molecules expressed specifically at the surface of porcine γδ-T cells. Immunology, 115(2), 189–196. https://doi.org/10.1111/j.1365-2567.2005.02137.x