Scientists cannot take a picture of an atom. This is because visible light consists of waves so much larger than atoms that the two do not interact. Even supposed pictures of atoms, like this particularly famous one by British physicist David Nadlinger, show the radiation emitting from the atom – not the atom itself. So how else are we supposed to observe what happens at the atomic scale?

Vibration is an important key to link the quantum world with our own. Whereas visual observation is difficult on ultra-small scales, vibration still applies as a valid way to detect information. It’s like how you may be able to feel a splinter in your finger even though you can’t see it. For convenience, physicists often describe vibrations – periodic movement of atoms – as “phonons” (Chandler, 2010). Though phonons may sound like fundamental particles like protons or electrons, they aren’t. Phonons are just a way to describe complicated interactions between other fundamental particles into an easy-to-work-with particle form (Lewton, 2021). What we call heat is the same sort of approximation: it is an aggregate, easier way to describe the energy of the innumerable atoms in an object colliding like hundreds of billiard balls.
In Nature Physics, a team of researchers led by Caltech Professor Alkim Bozkurt recently created a device capable of reliably transforming phonons into detectable electromagnetic waves (2023). In their paper, published June of 2023, they describe an electrical circuit whose outputs vary with vibration.
Fundamental to the researchers’ design is a capacitor. Capacitors are circuit components that store electrical charges, i.e. electrons, on two parallel conducting plates separated by a small distance on their bodies. Capacitors are defined by their capacitance, which is the ability of a given component to hold charge. The key to Bozkurt et al.’s device is that capacitance depends on the distance between its parallel plates (2023). When the distance between the plates increases, capacitance decreases, and vice versa.

In the device described in the paper, Bozkurt et al. take advantage of capacitors’ properties to transmit photons (2023). The researchers connected the parallel plates of a calculator to a vibrating crystal lattice. When the lattice moves, the parallel plates – and thus capacitance – change accordingly. Oscillating capacitance isn’t detectable on its own. However, voltage is a function of capacitance, and capacitance is the function of vibration. By hooking the capacitor up to a circuit whose output changes with voltage, we can observe the information carried by the phonons.
By hooking up the capacitor to a small transmitting circuit, the researchers can ascertain the phonons’ properties (Bozkurt et al., 2023). In operation, the wave transmitter must be tuned until the interaction between the moving capacitor and the transmitter creates a peak of detectable voltage in the transmitted wave. This peak occurs where the phonon frequency matches that of the transmitted microwave.
Say, for example, you have a crystal that vibrates at an unknown frequency 𝑓 that is greater than a millions of times per second. To find 𝑓, we attach the new device to the crystal. Then, once it begins vibrating, you must tune the transmitter’s electromagnetic wave output frequency to match the unknown 𝑓. When a peak of detectable voltage in the transmitted microwaves is observed, the output microwave frequency will equal to the unknown 𝑓. Thus, by measuring the microwaves, you will have indirectly measured the frequency of the vibrating crystal.

Such a device has practical applications well beyond measurement. The device can be used in parts of quantum memory units, interfacing with parts of quantum computers. It could effectively record the characteristic changes of a quantum system and then restore that state accordingly – something optical observing at such a small scale cannot achieve. Still, the invention is not quite able to match the phonon frequency as alternatives. Its outputs sometimes do not agree with the known vibration of the crystal. The paper cites possible next steps – including altering the transmitter – to increase its agreement with the known vibration of the phonons in testing (Bozkurt et al., 2023).
Still, this development is remarkable. The Caltech researchers produced a device that detects phonons at speeds of a billion times a second, orders of magnitude more sensitive than similar previous devices, with substantially improved reliability (Bozkurt et al., 2023). Other similar approaches rely on hard-to-create materials and suffer from short lifetimes at small scales.
Works Cited:
Bozkurt, A., Zhao, H., Joshi, C., LeDuc, H. G., Day, P. K., & Mirhosseini, M. (2022). A quantum electromechanical interface for long-lived phonons. Nature Physics, 19(September 2023), 1326–1332.
Brean, J. (2018, February 14). Stunning image of a single strontium atom wins British photography prize. https://nationalpost.com/news/world/stunning-image-of-a-single-strontium-atom
Chandler, D. L. (2010, July 8). Explained: Phonons. MIT News. https://news.mit.edu/2010/explained-phonons-0706
Lewton, T. (2021, March 24). The Near-Magical Mystery of Quasiparticles. Quantamagazine. https://www.quantamagazine.org/the-near-magical-mystery-of-quasiparticles-20210324/
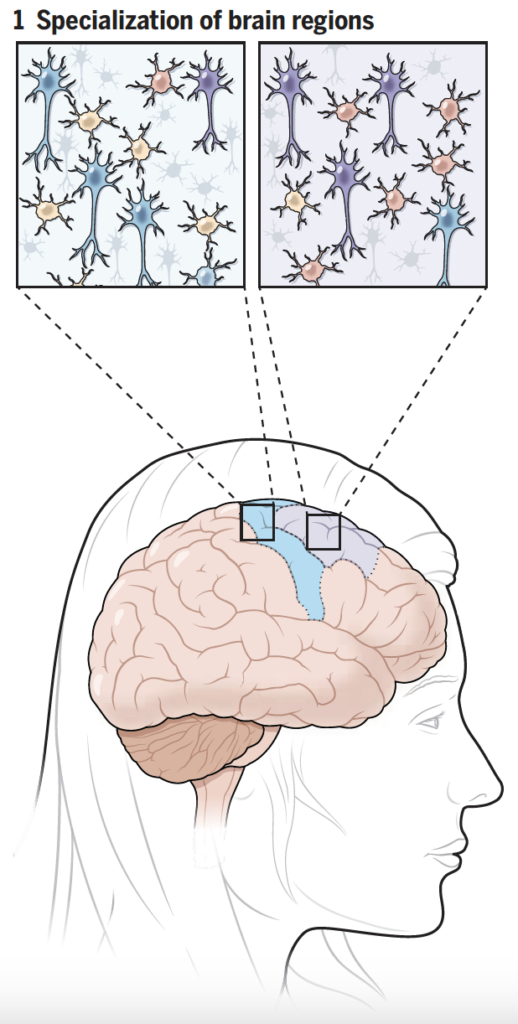
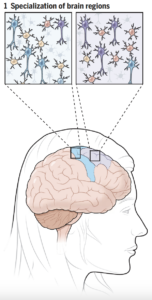 Comparisons of cell composition in regions across the brain resulted in findings from researchers under Siletti from the University of North Carolina at Chapel Hill and Jorstad from Harvard University. The two groups found that rather than mainly having different types of cells in different parts of the brain, some different parts of the brain shared the same cells but had different proportions of these cells. There were some exceptions, such as inhibitory neurons in the primary visual cortex, although the explanation of this finding is unclear. Such results change the understanding of evolutionary diversity in that diversification does not depend heavily on having many different cell types, but rather on having varying proportions of cells with small differences.
Comparisons of cell composition in regions across the brain resulted in findings from researchers under Siletti from the University of North Carolina at Chapel Hill and Jorstad from Harvard University. The two groups found that rather than mainly having different types of cells in different parts of the brain, some different parts of the brain shared the same cells but had different proportions of these cells. There were some exceptions, such as inhibitory neurons in the primary visual cortex, although the explanation of this finding is unclear. Such results change the understanding of evolutionary diversity in that diversification does not depend heavily on having many different cell types, but rather on having varying proportions of cells with small differences.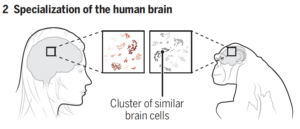
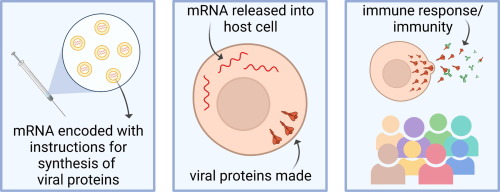
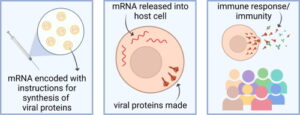

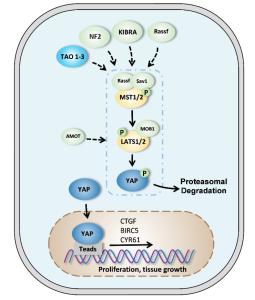
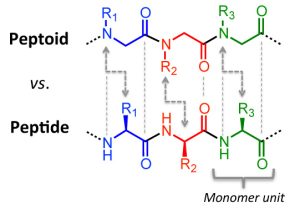
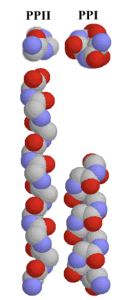
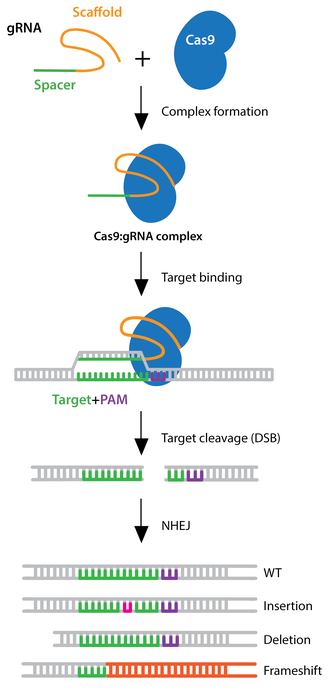


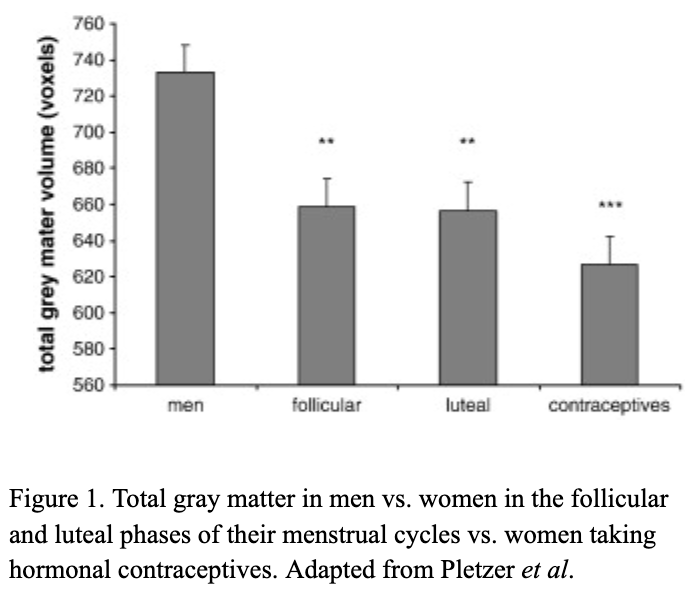
 There are millions of women taking steroids every day. But how is this possible? Are they just getting really buff? It feels like we always hear stories about how performance-enhancing drugs, namely steroids, are giving world-class athletes the boost they need to beat out their competition. But women across the globe are taking steroids every day as well, in the form of hormonal birth control. Despite their widespread use, side effects of hormonal contraceptives are largely unstudied, or have been until recently. In the last ten years, several studies have come out about the effect of taking a daily dose of steroids on women’s brains and mental health, which until now has been a severely neglected area where lack of knowledge affects millions of people worldwide.
There are millions of women taking steroids every day. But how is this possible? Are they just getting really buff? It feels like we always hear stories about how performance-enhancing drugs, namely steroids, are giving world-class athletes the boost they need to beat out their competition. But women across the globe are taking steroids every day as well, in the form of hormonal birth control. Despite their widespread use, side effects of hormonal contraceptives are largely unstudied, or have been until recently. In the last ten years, several studies have come out about the effect of taking a daily dose of steroids on women’s brains and mental health, which until now has been a severely neglected area where lack of knowledge affects millions of people worldwide. 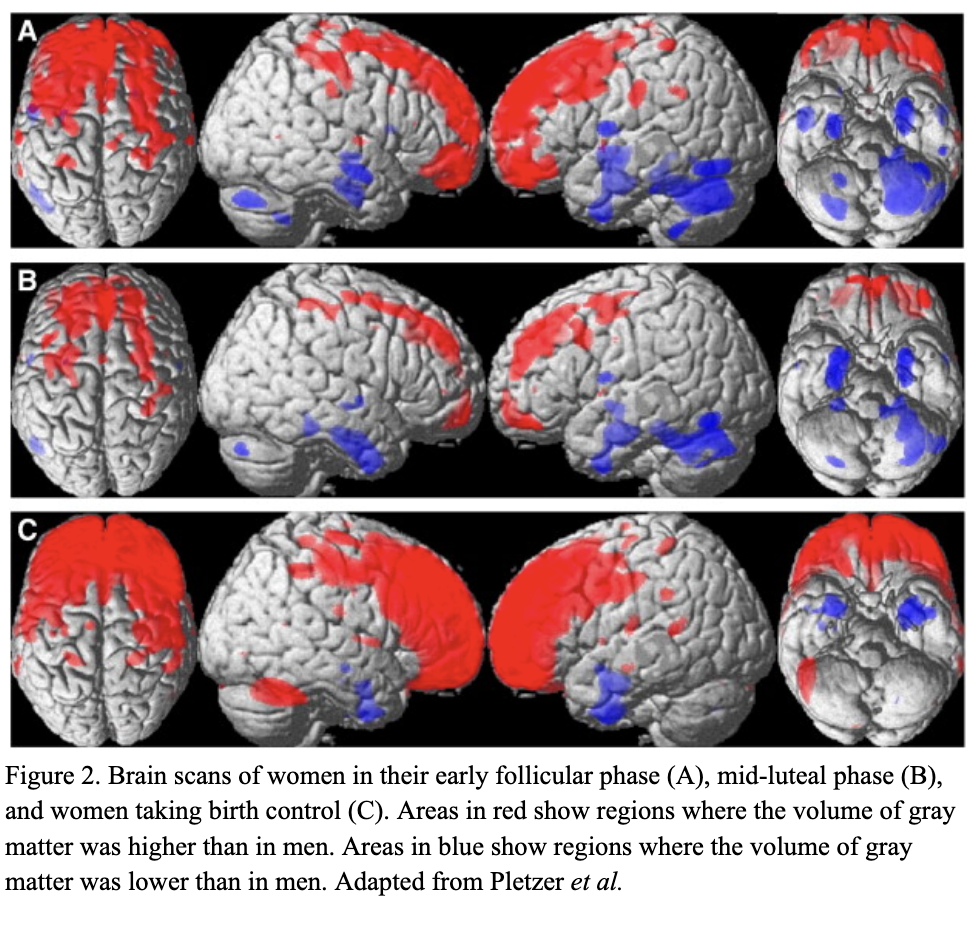 Thes
Thes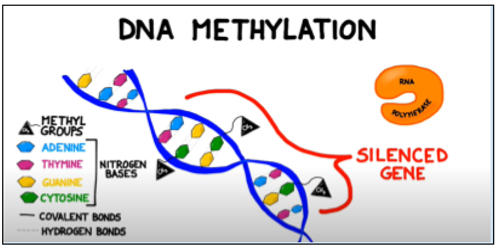

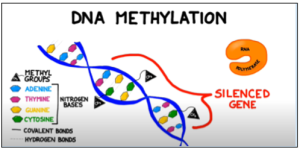 Figure 1: Methyl groups attached to cytosine bases in a gene block the enzyme RNA polymerase from binding to the promoter region of a gene, preventing transcription. Adapted from BOGOBiology (2017)
Figure 1: Methyl groups attached to cytosine bases in a gene block the enzyme RNA polymerase from binding to the promoter region of a gene, preventing transcription. Adapted from BOGOBiology (2017)