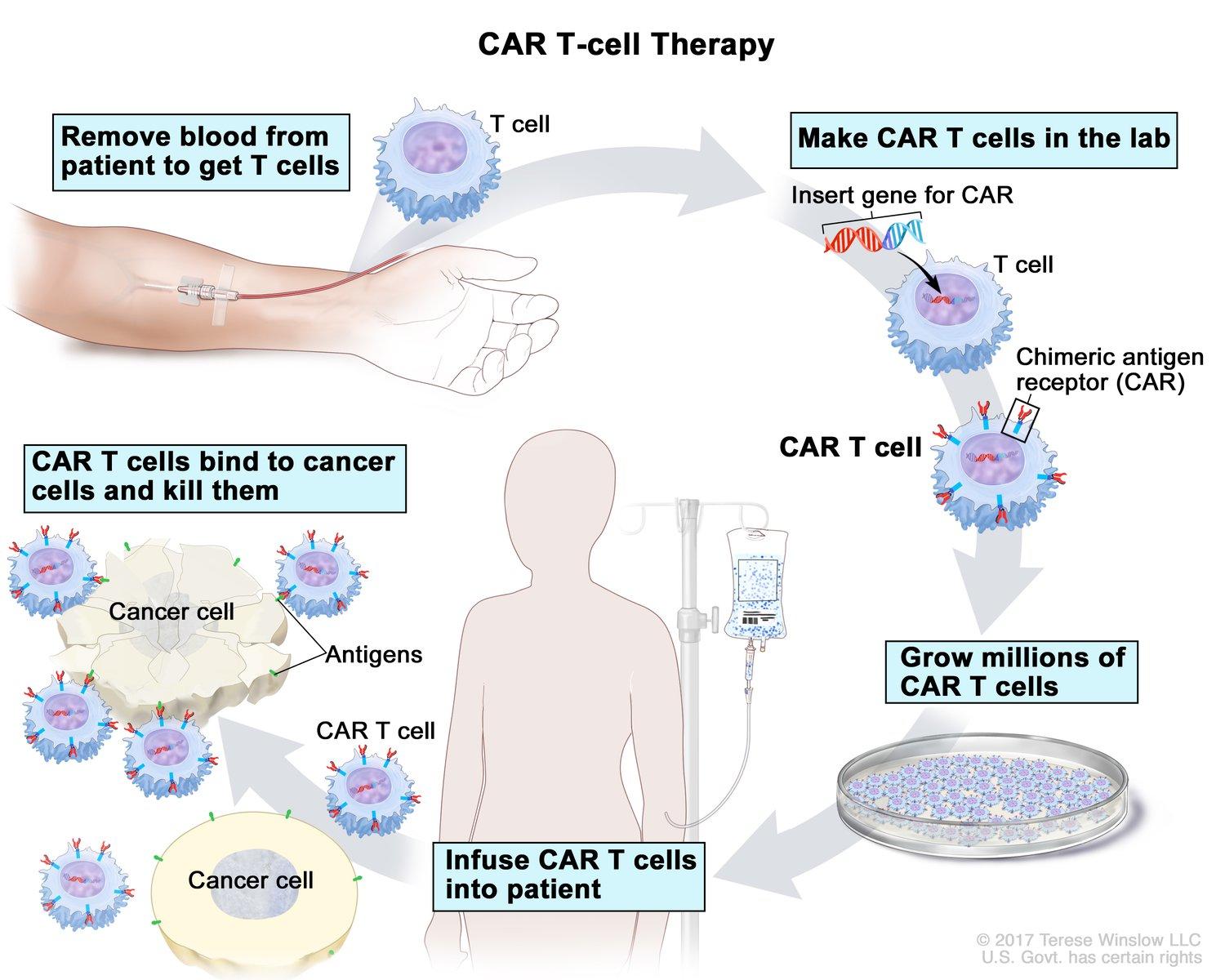
The prevalence of allergic diseases increased globally following the 1960s. Between 1982 and 1997, the prevalence of asthma and hay fever in Australian schoolchildren rose from 12.9 to 38.6% and 22.5 to 44.0%, respectively (Downs et al., 2001). Similar trends are observed globally (Thomsen, 2015; Turke, 2017). Allergic asthma occurs in about 12 million individuals in the U.S. and its prevalence continues to rise (Gutowska-Ślesik et al., 2023; GenenTech, n.d.). The immune system is the body’s form of defense against pathogens like viruses and bacteria, and is also where allergies begin. When the immune system regularly overreacts to a harmless substance, one is said to have an allergic disease (Allergies and the Immune System, 2021). Allergies are the subject of many immunological studies due to their health effects. Asthma, for example, is characterized by minor or life-threatening inflammation in the airways.
Theories have surfaced in order to explain the dramatic increase in allergic diseases. One leading theory is the Hygiene Hypothesis. The hypothesis claims that lack of exposure to certain microbial species, like bacteria, is important for the proper development of our immune system (Bloomfield et al., 2006). Therefore, researchers have investigated the mechanisms by which allergic disease, particularly asthma, is deterred by these species. A 2023 study by Yao and colleagues focuses on PepN—a bacterial protein that has shown promise in previous studies—to uncover how the immune system changes when exposed to allergy-reducing disease.
Alveolar macrophages (AMs) are a kind of macrophage found in the lungs that substantially influence the development of asthma (macrophages are a type of immune cell). AMs produce signalers that either encourage or inhibit inflammation in the lungs, which makes them targets for asthma treatments. In fact, AMs, when activated, undergo reprogramming that transforms them into a pro-infammatory or anti-infammatory macrophage (referred to as a CD11chigh macrophage). Yao and colleagues believe this process to be the theoretical foundation of the Hygiene Hypothesis, suggesting that asthma can be treated or prevented by deliberately transforming macrophages to be anti-inflammatory.
Yao and colleagues induced allergic asthma in mice through the use of intranasally injected allergens. To serve as a baseline, the control group was given no further treatment; in the experimental group, mice were exposed to bacterial protein PepN multiple times before and after being inflicted with asthma. Yao and colleagues then dissected the mice, investigating the CD11chigh macrophages and other forces at play.
After comparing the control group of mice to the experimental group, PepN was found to recruit macrophages from the bone marrow into the respiratory tract and transform them to be anti-inflammatory through changes in the macrophages’ metabolism. PepN also encouraged the proliferation of already existing CD11chigh macrophages that reside in the lungs. These forces culminated in a protective effect against allergic asthma (see fig. 1).

For the future, Yao and colleagues believe more research is necessary to determine other mechanisms of the Hygiene Hypothesis. Though there are limitations to their current study, Yao and colleagues provide a new idea for the prevention and treatment of allergic asthma: targeting CD11chigh macrophages to combat asthmatic inflammation.
References
Allergies and the immune system. (2021, August 8). Johns Hopkins Medicine. https://www.hopkinsmedicine.org/health/conditions-and-diseases/allergies-and-the-immune-system
Bloomfield, S. F., Stanwell‐Smith, R., Crevel, R., & Pickup, J. C. (2006). Too clean, or not too clean: the Hygiene Hypothesis and home hygiene. Clinical & Experimental Allergy/Clinical and Experimental Allergy, 36(4), 402–425. https://doi.org/10.1111/j.1365-2222.2006.02463.x
Downs, S. H., Marks, G. B., Sporik, R., Belosouva, E. G., Car, N., & Peat, J. K. (2001). Continued increase in the prevalence of asthma and atopy. Archives of Disease in Childhood, 84(1), 20–23. https://doi.org/10.1136/adc.84.1.20
GenenTech: Asthma Statistics. (n.d.). https://www.gene.com/patients/disease-education/asthma-statistics#:~:text=Prevalence,asthma%20sufferers%20in%20the%20U.S
Gutowska-Ślesik, J., Samoliński, B., & Krzych‐Fałta, E. (2023). The increase in allergic conditions based on a review of literature. Postępy Dermatologii I Alergologii, 40(1), 1–7. https://doi.org/10.5114/ada.2022.119009
Thomsen, S. F. (2015). Epidemiology and natural history of atopic diseases. European Clinical Respiratory Journal, 2(1), 24642. https://doi.org/10.3402/ecrj.v2.24642
Turke, P. W. (2017). Childhood food allergies. Evolution, Medicine and Public Health, 2017(1), 154–160. https://doi.org/10.1093/emph/eox014
Yao, S., Weng, D., Wang, Y., Zhang, Y., Huang, Q., Wu, K., Li, H., Zhang, X., Yin, Y., & Xu, W. (2023). The preprogrammed anti-inflammatory phenotypes of CD11chigh macrophages by Streptococcus pneumoniae aminopeptidase N safeguard from allergic asthma. Journal of Translational Medicine, 21(1). https://doi.org/10.1186/s12967-023-04768-2