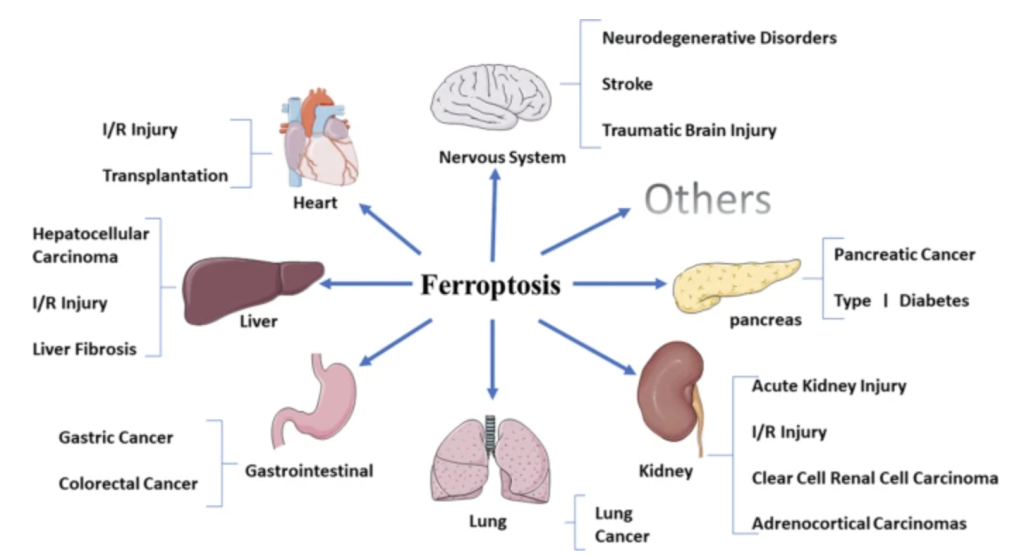
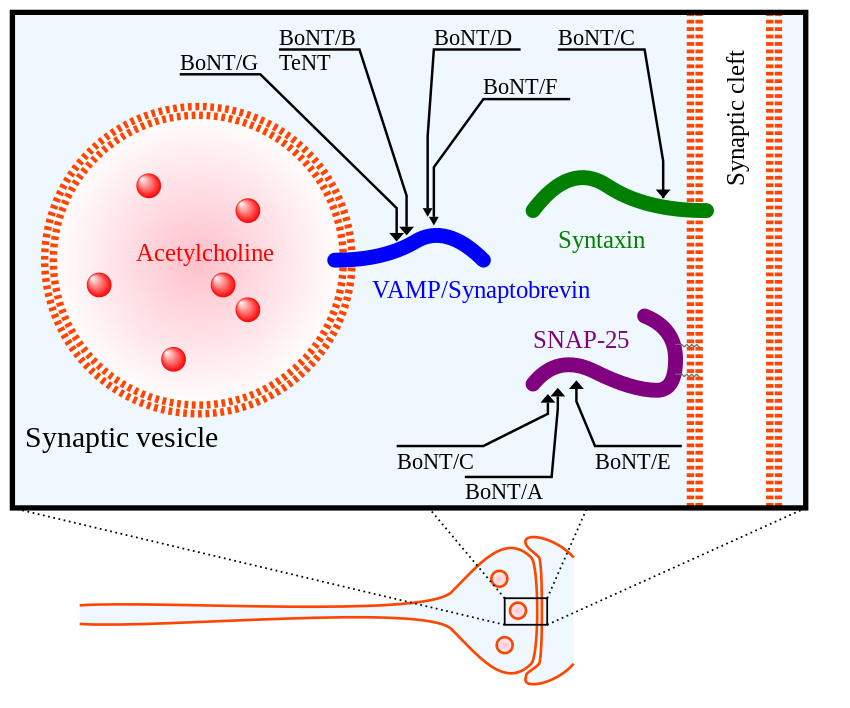
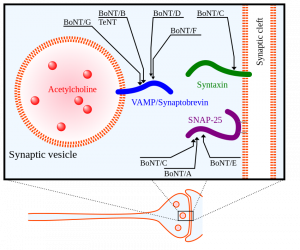
Did you know that horseshoe crabs have incredible immune systems? In fact, horseshoe crabs have the best immune systems out of all living invertebrates. Their secret? Blood. Horseshoe crab blood is very simple in composition, with only a single cell type in general circulation (the granular amebocyte) and only three proteins in the plasma of the blood (hemocyanin, C-reactive proteins, and a2-macroglobulin) (Armstrong et al., 2008). These proteins contribute to the horseshoe crab’s blood clotting system, protecting them from infection. Horseshoe crab blood has been found to be very sensitive to bacterial endotoxins found in illness-causing Gram-negative bacteria (Protecting Health). When horseshoe crab blood cells come into contact with bacterial endotoxin, they clot around it, preventing the bacterium from invading nearby cells (Natural History Museum 2020).
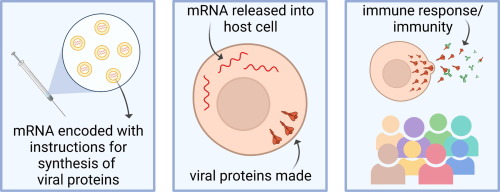
With the rise of vaccine development, especially in the case of the Covid-19 pandemic, horseshoe crab blood plays an essential role in testing the safety of vaccines due to its endotoxin-detection properties. Additionally, large volumes of horseshoe crab blood can be collected easily, making it a convenient blood source (Armstrong et al., 2008). Despite all the beneficial applications of horseshoe crab blood, horseshoe crab bleeding leaves thousands of horseshoe crabs dead annually, causing their populations to be in decline (Maloney et al., 2018). A 2018 study has promoted a synthetic alternative to horseshoe crab blood, recombinant Factor C (rFC), and proven its efficacy in bacterial endotoxin detection. The use of rFC in vaccine development can eliminate the need for the use of actual horseshoe crab blood, sparing the horseshoe crab and promoting the conservation of this endangered species.
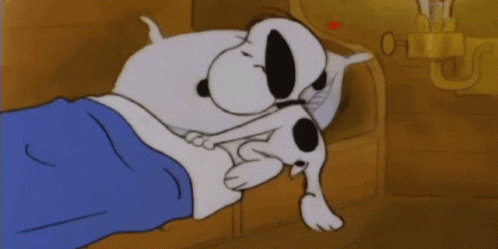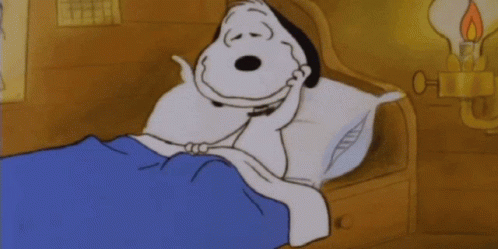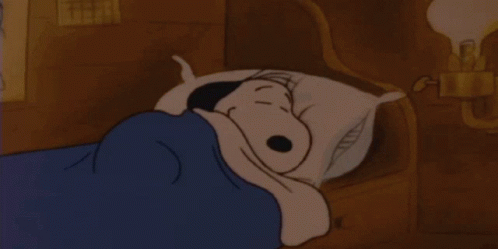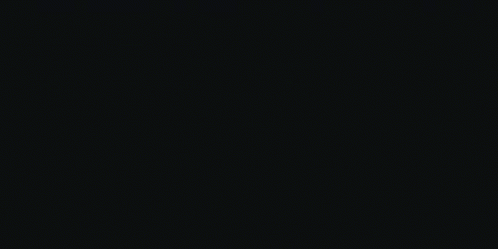
Typically, horseshoe crab blood is collected by the direct puncture of the heart under sterile conditions that minimize contamination by bacterial endotoxins (Figure 1). A large horseshoe crab can produce between 200 – 400 mL of blood, and the blood clotting system can be studied microscopically. The limitation of contamination by bacterial endotoxins is extremely important in the blood collection process, because cell clotting will compromise the effectiveness of the blood for its intended use of developing vaccines. Only undamaged horseshoe crabs are selected for blood collection, and the animal is bled by the insertion of a needle into the heart through the outer hinge joint of the horseshoe crab (Figure 2). The animal is then squeezed gently so that as much blood as possible can be deposited into the collection tube (Armstrong et al., 2008).

Figure 1: Horseshoe crab bleeding on a larger scale, with precautions taken to ensure sanitary, endotoxin-free conditions for blood collection.
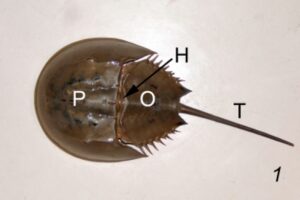
Figure 2: The three major components of the body of a horseshoe crab, including the prosoma (P), the opisthosoma (O), and the telson (T). The hinge (H) is where the prosoma meets the opisthosoma, and that is where the needle is inserted for blood collection.
Following collection, horseshoe crab blood is ready for use in endotoxin detection (Armstrong et al., 2008). For example, in vaccine development, a Limulus amebocyte lysate (LAL) test detects the levels of clotting in horseshoe crab blood when it comes into contact with different vaccines. Horseshoe crab blood is very precise with detecting even small traces of endotoxin, making it an effective tool to identify small quantities of endotoxin present in potential vaccines (Protecting Health).
Although horseshoe crab blood is effective in its ability to detect endotoxins, recombinant Factor C (rFC) can do the same job in a way that is more ecologically sustainable. Initially, rFC was discovered by scientists at the National University of Singapore, allowing them to visualize endotoxin detection using animal-free technology. Every year over 500,000 horseshoe crabs are captured and as much as ⅓ of their blood is drained, contributing to high mortality rates. On top of this, around 13% of the horseshoe crabs bled are later sold for bait, resulting in nearly 130,000 horseshoe crab victims to the biomedical industry. A 2018 study confirmed that the biomedical industry could reduce their use of horseshoe crab blood by nearly 90% if they were to employ rFC as a synthetic alternative for endotoxin detection processes. The 2018 study reviews multiple studies that show how rFC is just as effective as actual horseshoe crab blood in endotoxin detection, as rFC has been able to demonstrate the same high rate and sensitivity as horseshoe crab blood in detecting small amounts of endotoxin in a wide range of chemical structures. When endotoxin binds to a synthetic rFC molecule, it causes the rFC to fluoresce directly proportional to the concentration of endotoxin in a substance. rFC has even been able to demonstrate a higher rate of specificity for endotoxin detection (compared to horseshoe crab blood) in some studies (Maloney et al., 2018).
The most important next step of this research is to get synthetic rFC into the hands of the biomedical industry. Exposure to endotoxin can cause serious illness, making endotoxin detection for vaccines an essential part of the vaccine development process. Even though there is ample evidence that rFC is equivalent to or better than horseshoe crab blood at detecting bacterial endotoxin, there are still limitations to the usage of rFC, as it is difficult for the biomedical industry to adopt new technologies quickly. Endotoxin detection testing is very highly regulated, so many pharmaceutical manufacturers may be hesitant to employ new detection technologies, as they may want to stick to traditional methods instead (Maloney et al., 2018). Despite these limitations, in order to progress towards horseshoe crab conservation, rFC should be produced and employed on a large scale so that the biomedical industry will no longer be solely reliant on the exploitation of horseshoe crabs for bacterial endotoxin detection.
Literature Cited
1. Armstrong, P., Conrad, M. Blood Collection from the American Horseshoe Crab, Limulus Polyphemus. J. Vis. Exp. (20), e958, doi:10.3791/958 (2008).
2. “Horseshoe Crab Blood: The Miracle Vaccine Ingredient That’s Saved Millions of Lives.” Www.nhm.ac.uk, www.nhm.ac.uk/discover/horseshoe-crab-blood-miracle-vaccine-ingredient.html#:~:text=Horseshoe%20crab%20blood%20is%20bright.
3. Maloney T, Phelan R, Simmons N. Saving the horseshoe crab: A synthetic alternative to horseshoe crab blood for endotoxin detection. PLoS Biol. 2018 Oct 12;16(10):e2006607. doi: 10.1371/journal.pbio.2006607. PMID: 30312293; PMCID: PMC6200278.
4. “Protecting Health.” Www.horseshoecrab.org, www.horseshoecrab.org/med/health.html. Accessed 12 Nov. 2023.