Jellyfish are just one species within the phylum cnidaria. A phylum is a broad level of taxonomic classification that includes many different species, with cnidaria additionally including coral and anemones. Cnidaria provides comparative neuroscience information due to the simple behaviors that the species within the phylum exhibit. Despite their shared phylum that creates nerve cells with similar properties, the species have dramatically different nervous systems, allowing for unique perspectives on the diversity, origins, and evolution of neural systems within species (Cunningham et al., 2024). Comparative neuroscience information is the study of nervous systems across a variety of animal species. Through this research, the evolutionary changes in the brain’s structure can be examined, allowing scientists to see how differences in nervous systems shape certain behaviors (Miller et al., 2019). Neuroscience researchers can use an all-optical interrogation, in which they study and manipulate neural systems using light, upon these species, allowing them to image and photograph the neuronal networks in the creature for further examination.

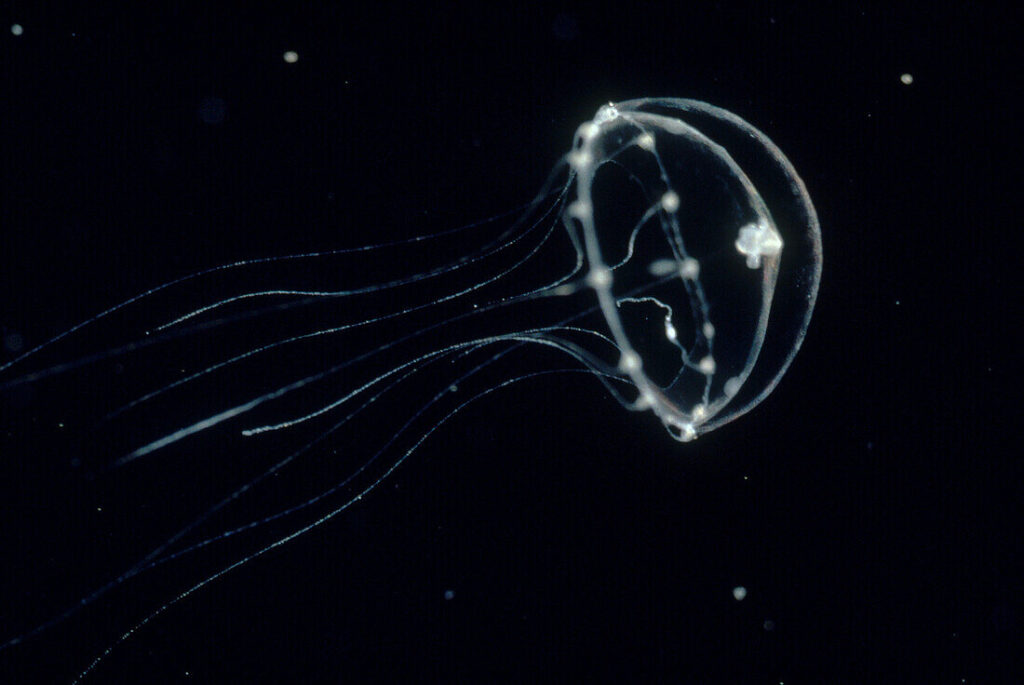
Fig 1. Photo of Clytia hemisphaerica (Clytia Hemisphaerica Medusa – 13673149 ❘ Science Photo Library, n.d.)
Jellyfish are major contributors to ocean ecosystems. Their reproductive, foraging, and defensive behaviors all uniquely impact the ecosystem at large. What is notable about jellyfish, however, is that these behaviors are shaped out of decentralized, regenerative nervous systems. Rather than the creature being controlled by the neurons in its brain, the jellyfish’s neurons are spread throughout the body (“Thinking without a Centralized Brain,” n.d.). This allows the various parts of its body to have a role in controlling and processing information. Additionally, the nervous system itself has the ability to repair and restore itself, allowing damaged nerves to be replaced by new ones (Gaskill, 2018).
Jellyfish are the most complicated species of Cnidaria, due to various behaviors that demonstrate their higher level functioning compared to other species in the phylum. They have the ability to move in 3-dimensions, capture and consume other creatures, and the ability to escape from predators and other potential threats. Notably, jellyfish also exhibit courtship behaviors and sleep states, despite lacking a central brain. These behaviors are due to their sensory structures, made up by two nervous systems: one which controls their swimming and another that controls all other behaviors. The jellyfish’s nervous systems can respond to each other, despite the lack of a central controller (Cunningham et al., 2024).
A recent scientific investigation conducted at Caltech by Anderson et al. sought to explore how the jellyfish can be used to conduct neuroscience research. Clytia hemisphaerica is a species of jellyfish that has recently been adopted into a genetic neuroscience model. It has previously been used as a model to study evolution, embryology, regeneration, and other fields. This species of jellyfish is a particularly useful model for neuroscience research because its genome is already sequenced and assembled from the birth of the creature, with whole-animal single-celled Ribonucleic Acid (RNA) sequences formed within the species. Rather than using multiple animals to sequence the RNA, Clytia hemisphaerica has the capability to provide the necessary amounts of cells needed to be examined. Using multiplexed single-cell RNA sequencing, in which individual animals were indexed and pooled from control and perturbation conditions into a single sequencing run (Chari et al., 2021). Clytia hemisphaerica is the only jellyfish whose RNA sequences are being used to rapidly develop genomic tools. These tools can be tested and utilized by researchers, allowing them to explore brain function and neurological disorders through this model (Cunningham et al., 2024).
The last common ancestor of Clytia hemisphaerica was a hydrozoan jellyfish, which are able to perform specific behaviors even if certain body parts are detached from the body. Hydrozoan jellyfish, notably, have the ability to cycle back and forth between various stages of their life–allowing them to live for large expanses of time. When the body parts exist in an intact organism, they also have the ability to perform more complex behaviors. These include different feeding behaviors and mechanisms. The swimming behavior of the Clytia hemisphaerica also reveals key information about the neuromechanics behind different behaviors of jellyfish. As an example, although jellyfish spend a majority of their lives swimming, there are periods where they may start and stop. These periods are only exhibited when there is food passing or defensive behaviors are exhibited. When these behaviors are examined, neuroscientists can ponder and develop further conclusions about multi-sensory integration, motor control, and the mechanisms that underlie behavioral states (Cunningham et al., 2024). 
Fig 2. The Evolution, Life Cycle, and Genetic Tools of Clytia hemisphaerica (Cunningham et al., 2024)
Through using neural population imaging, in which researchers have the ability to monitor large groups of neurons through calcium and voltage imaging, on the whole-organism scale through the Clytia hemisphaerica, emergent properties of function networks can be uncovered (Zhu et al., 2022). Without this model, scientists would have to use traditional single-cell unit recordings, requiring using fine tools just to see the individual activity of a single neutron, or anatomical studies, which would not provide the same amount of potential discoveries that new techniques with Clytia hemisphaerica provide. Through using this species as a model, researchers can uncover more knowledge and data about nervous system evolution and function, particularly for neural regeneration.
Neural regeneration is particularly important in the treatment of injury and disease in the nervous system. It aids in cognitive recovery following neurodegeneration, helping rebuild neurons and nervous tissue (Steward et al., 2013). Through neural regeneration, the nervous system may regain its functions, allowing for betterment of quality of life. By continuing to examine species capable of neural regeneration, we may learn to apply this to the human nervous system, allowing us to move forward in curing traumatic brain injuries and degeneration of the brain and its abilities (Neuroregeneration – an Overview | Sciencedirect Topics, n.d.).
References:
Chari, T., Weissbourd, B., Gehring, J., Ferraioli, A., Leclère, L., Herl, M., Gao, F., Chevalier, S., Copley, R. R., Houliston, E., Anderson, D. J., & Pachter, L. (2021). Whole animal multiplexed single-cell rna-seq reveals plasticity of clytia medusa cell types. bioRxiv. https://doi.org/10.1101/2021.01.22.427844
Cunningham, K., Anderson, D. J., & Weissbourd, B. (2024). Jellyfish for the study of nervous system evolution and function. Current Opinion in Neurobiology, 88, 102903. https://doi.org/10.1016/j.conb.2024.102903
Gaskill, M. (2018, November 20). No brain? For jellyfish, no problem | blog | nature | pbs. Nature. https://www.pbs.org/wnet/nature/blog/no-brain-for-jellyfish-no-problem/
Miller, C. T., Hale, M. E., Okano, H., Okabe, S., & Mitra, P. (2019). Comparative principles for next-generation neuroscience. Frontiers in Behavioral Neuroscience, 13. https://doi.org/10.3389/fnbeh.2019.00012
Neuroregeneration—An overview | sciencedirect topics. (n.d.). Retrieved April 27, 2025, from https://www.sciencedirect.com/topics/neuroscience/neuroregeneration
Steward, M. M., Sridhar, A., & Meyer, J. S. (2013). Neural regeneration. Current Topics in Microbiology and Immunology, 367, 163–191. https://doi.org/10.1007/82_2012_302
Thinking without a centralized brain: The intelligence of the octopus. (n.d.). WHYY. Retrieved April 27, 2025, from https://whyy.org/segments/thinking-without-a-centralized-brain-the-intelligence-of-the-octopus/
Zhu, F., Grier, H. A., Tandon, R., Cai, C., Agarwal, A., Giovannucci, A., Kaufman, M. T., & Pandarinath, C. (2022). A deep learning framework for inference of single-trial neural population dynamics from calcium imaging with sub-frame temporal resolution. Nature Neuroscience, 25(12), 1724–1734. https://doi.org/10.1038/s41593-022-01189-0